12.1 The Iterative Overlap Peak Merging Procedure
We first introduced a strategy for iterative overlap peak merging in 2018. Other peak merging strategies suffer from a few key issues that we outline below.
12.1.1 Fixed-width vs Variable-width Peaks
We use 501-bp fixed-width peaks because they make downstream computation easier as peak length does not need to be normalized. Moreover, the vast majority of peaks in ATAC-seq are less than 501-bp wide. Using variable-width peaks also makes it difficult to merge peak calls from multiple samples. In general, we do not feel that the potential benefit derived from using variable-width peaks outweighs the costs. More broadly, most analyses are stable with respect to the peak set or peak style used.
Below, we use the same toy example of a few cell types with a few different peaks to illustrate the differences between these often used peak merging methods.
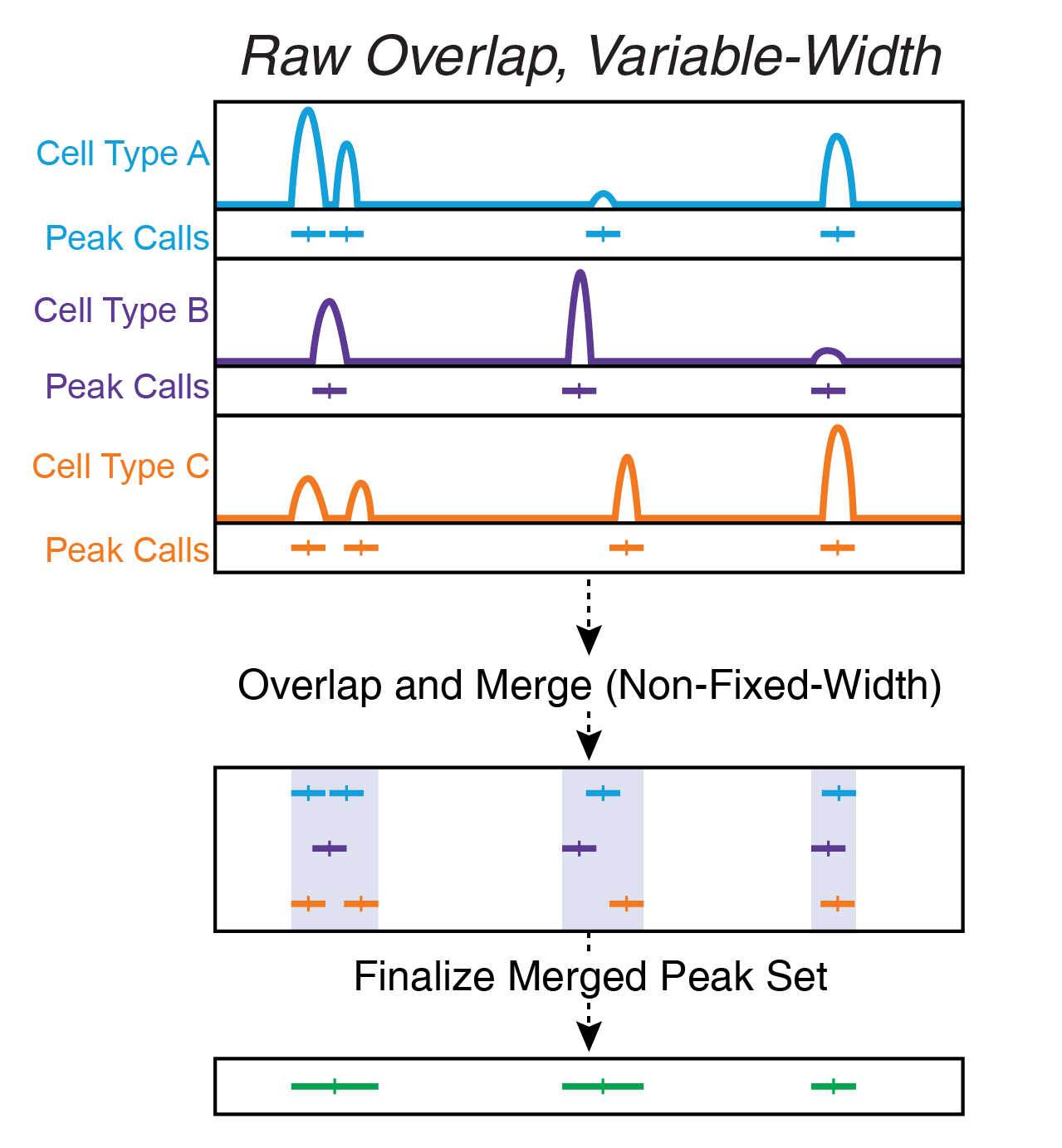
12.1.2 Raw Peak Overlap Using bedtools merge
Raw peak overlap involves taking any peaks that overlap each other and merging these into a single larger peak. In this scheme, daisy-chaining becomes a large problem because peaks that dont directly overlap each other get included in the same larger peak because they are bridged by a shared internal peak. Another problem with this type of approach is that, if you want to keep track of peak summits, you are forced to either pick a single new summit for each new merged peak or keep track of all of the summits that apply to each new merged peak. Typically, this type of peak merging approach is implemented using the bedtools merge command.
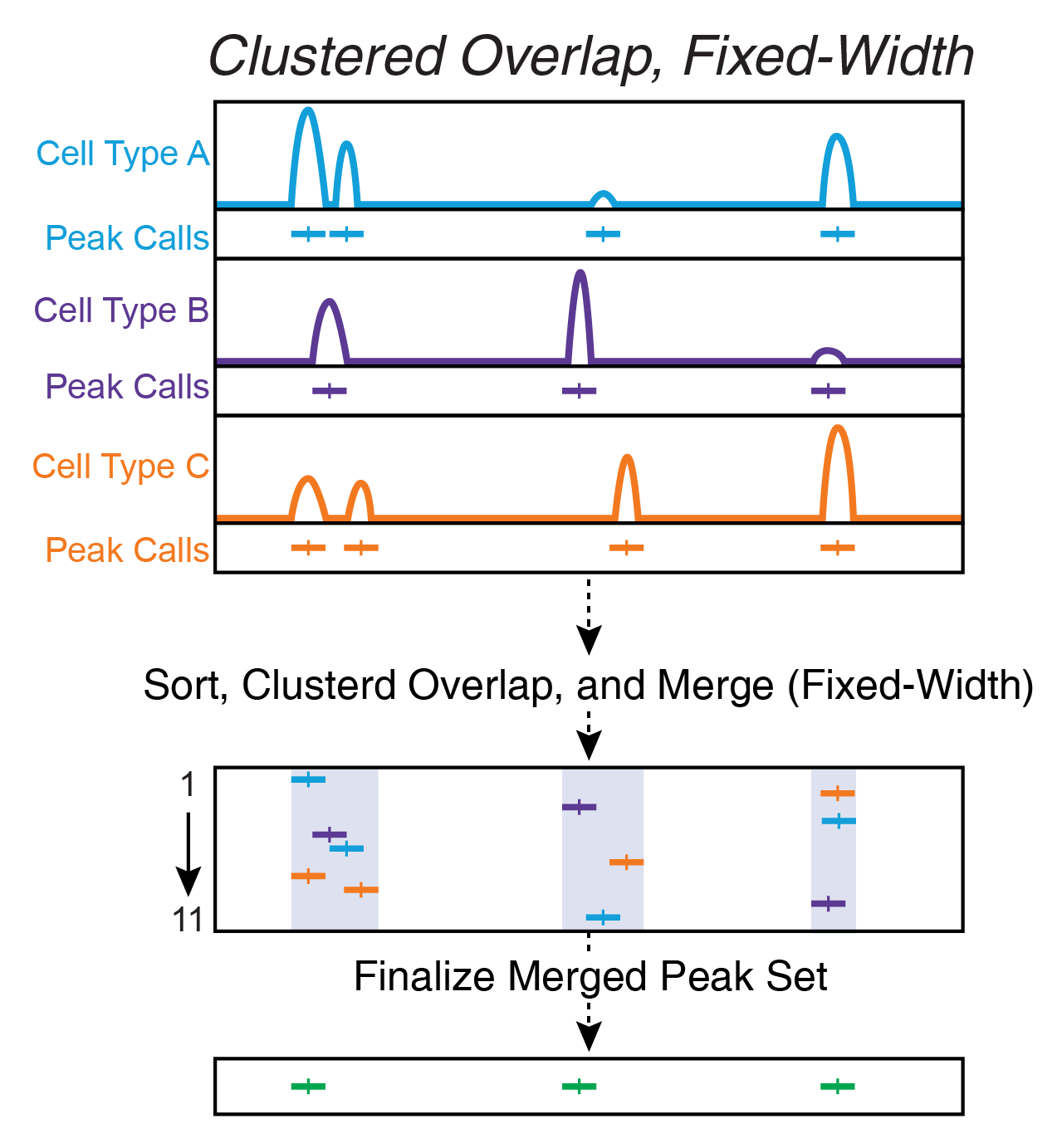
12.1.3 Clustered Overlap Using bedtools cluster
Clustered overlap takes peaks that cluster together and picks a single winner. This is often done by using the bedtools cluster command and then keeping the most significant peak in each cluster. In our experience, this ends up under-calling peaks and misses smaller peaks located nearby.
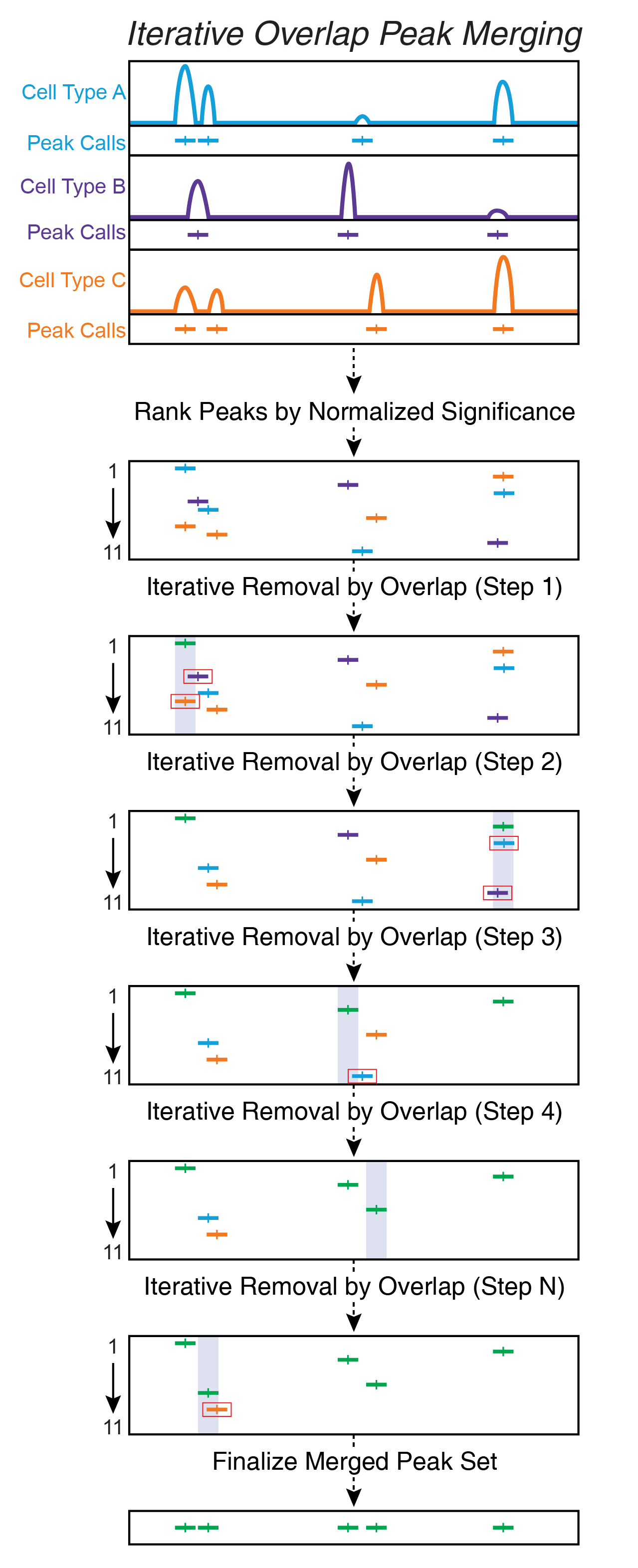
12.1.4 Iterative Overlap In ArchR
Iterative overlap removal avoids the issues mentioned above. Peaks are first ranked by their significance. The most significant peak is retained and any peak that directly overlaps with the most significant peak is removed from further analysis. Then, of the remaining peaks, this process is repeated until no more peaks exist. This avoids daisy-chaining and still allows for use of fixed-width peaks.
If you’ve come to this page looking to implement this iterative overlap peak merging procedure on bulk ATAC-seq data, this isn’t possible through ArchR but we host a separate repository that contains code to do this with bulk ATAC-seq.
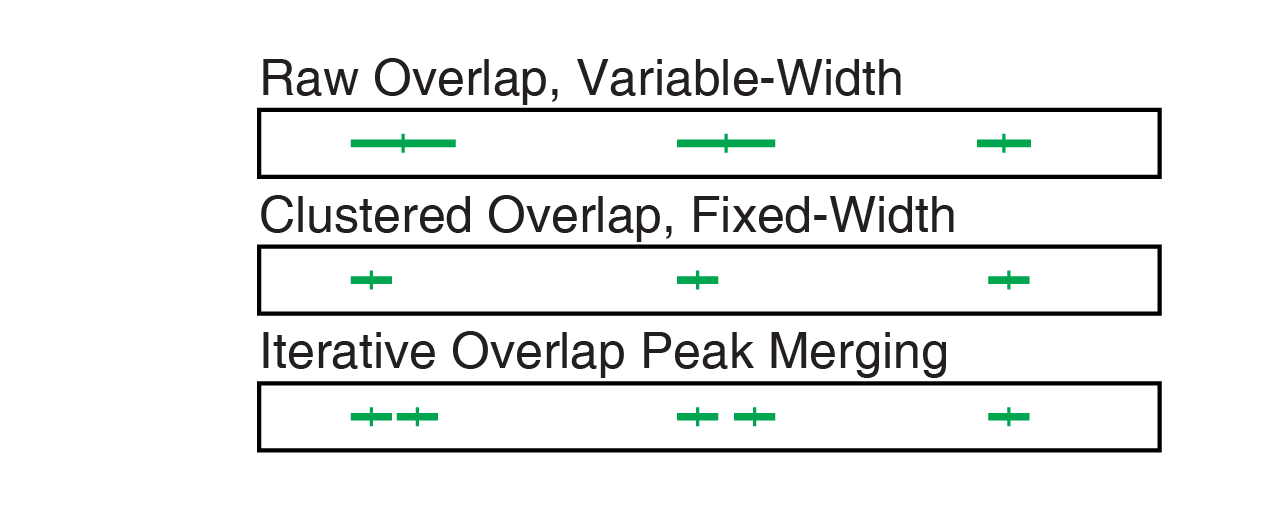
12.1.5 Comparison of Peak Calling Methods
Comparing the peak calls resulting from all of these methods directly shows clear differences in the final peak sets. It is our opinion that the iterative overlap peak merging process yields the best peak set with the fewest caveats.
12.1.6 So how does this all work in ArchR?
The iterative overlap peak merging procedure is performed in a tiered fashion to optimally preserve cell type-specific peaks.
Imagine a situation where you had 3 cell types, A, B, and C, and each cell type had 3 pseudo-bulk replicates. ArchR uses a function called addReproduciblePeakSet() to perform this iterative overlap peak merging procedure. First, ArchR would call peaks for each pseudo-bulk replicate individually. Then, ArchR would analyze all of the pseudo-bulk replicates from a single cell type together, performing the first iteration of iterative overlap removal. It is important to note that ArchR uses a normalized metric of significance for peaks to compare the significance of peaks called across different samples. This is because the reported MACS2 significance is proportional to the sequencing depth so peak significance is not immediately comparable across samples. After the first iteration of iterative overlap removal, ArchR checks to see the reproducibility of each peak across pseudo-bulk replicates and only keeps peaks that pass a threshold indicated by the reproducibility parameter. At the end of this process, we would have a single merged peak set for each of the 3 cell types, A, B, and C.
Then, we would repeat this procedure to merge the A, B, and C peak sets. To do this, we re-normalize the peak significance across the different cell types, and perform the iterative overlap removal. The final result of this is a single merged peak set of fixed-width peaks.