4.1 How does doublet identification work in ArchR?
Single-cell data generated on essentially any platform is susceptible to the presence of doublets. A doublet refers to a single droplet that received a single barcoded bead and more than one nucleus. This causes the reads from more than one cell to appear as a single cell. For 10x, the percentage of total “cells” that are actually doublets is proportional to the number of cells loaded into the reaction. Even at the lower levels of doublets that result from standard kit use, more than 5% of the data may come from doublets and this exerts substantial effects on clustering. This issue becomes particularly problematic in the context of developmental/trajectory data because doublets look like a mixture between two cell types and this can be confounded with intermediate cell types or cell states.
To predict which “cells” are actually doublets, we synthesize in silico doublets from the data by mixing the reads from thousands of combinations of individual cells. We then project these synthetic doublets into the UMAP embedding and identify their nearest neighbor. By iterating this procedure thousands of times, we can identify “cells” in our data whose signal looks very similar to synthetic doublets.
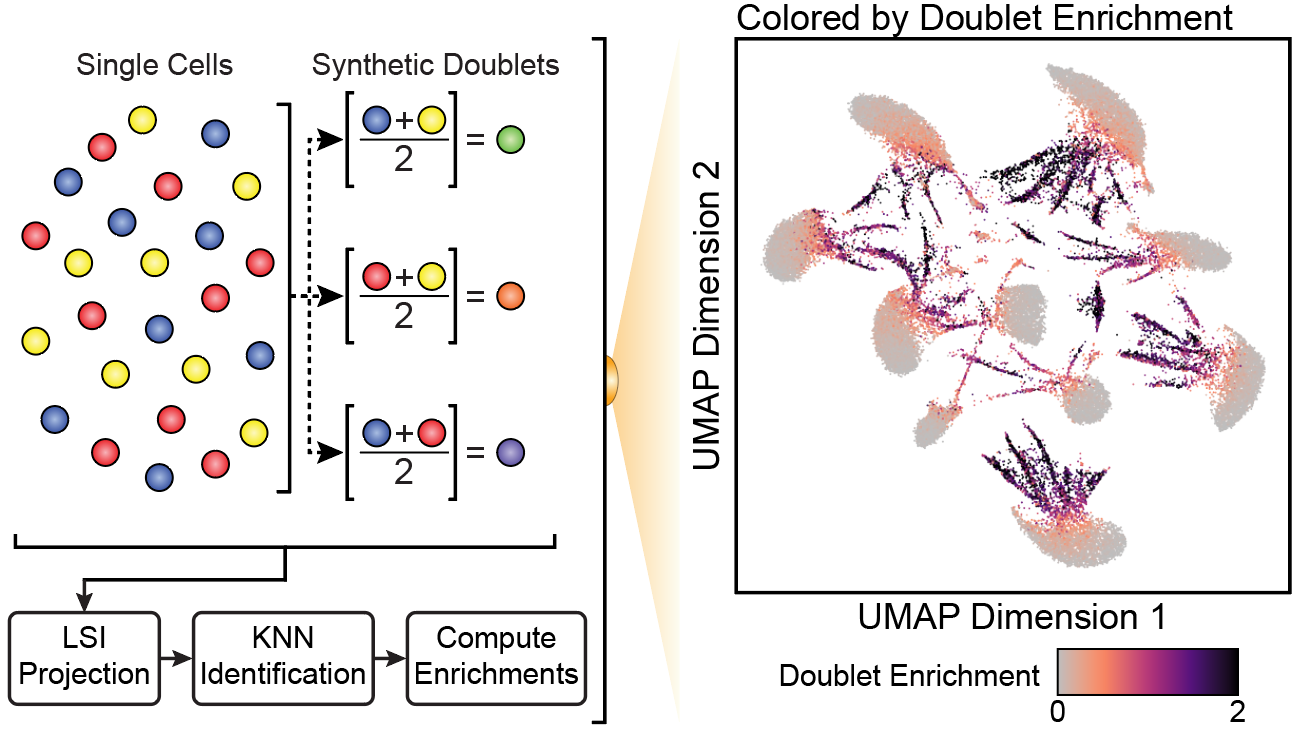
To develop and validate ArchR’s doublet identification, we generated scATAC-seq data from pooled mixtures of 10 genetically distinct cell lines. In scATAC-seq space, these 10 cell lines should form 10 distinct clusters but when we deliberately overload the 10x Genomics scATAC-seq reaction, targetting 25,000 cells per reaction, we end up with many doublets. We know these are doublets because we use demuxlet to identify droplets that contain genotypes from two different cell types.
This “ground truth” overlaps very strongly with the doublet predictions shown above, showing an area under the curve of the receiver opperating characteristic >0.90.
After we computationally remove these doublets with ArchR, the overall structure of our data changes dramatically and matches our expectation of 10 distinct cell types.