11.1 How Does ArchR Make Pseudo-bulk Replicates?
To create pseudo-bulk replicates, ArchR employs a tiered priority approach. The user specifies (i) the minimum and maximum number of replicates desired, (ii) the minimum and maximum number of cells per replicate, and (iii) the sampling ratio to use if a particular grouping lacks sufficient cells to make the desired replicates. For example, a sampling ratio of 0.8 means that cells can be sampled without replacement up to 80% of the total number of cells for each replicate (this will result in sampling with replacement across replicates). In this case, multiple replicates may contain some of the same cells but this is a necessary sacrifice if you would like to generate pseudo-bulk replicates from a cell group that lacks sufficient cells.
We process for pseudo-bulk replicate generation can be described by a decision tree as shown below.
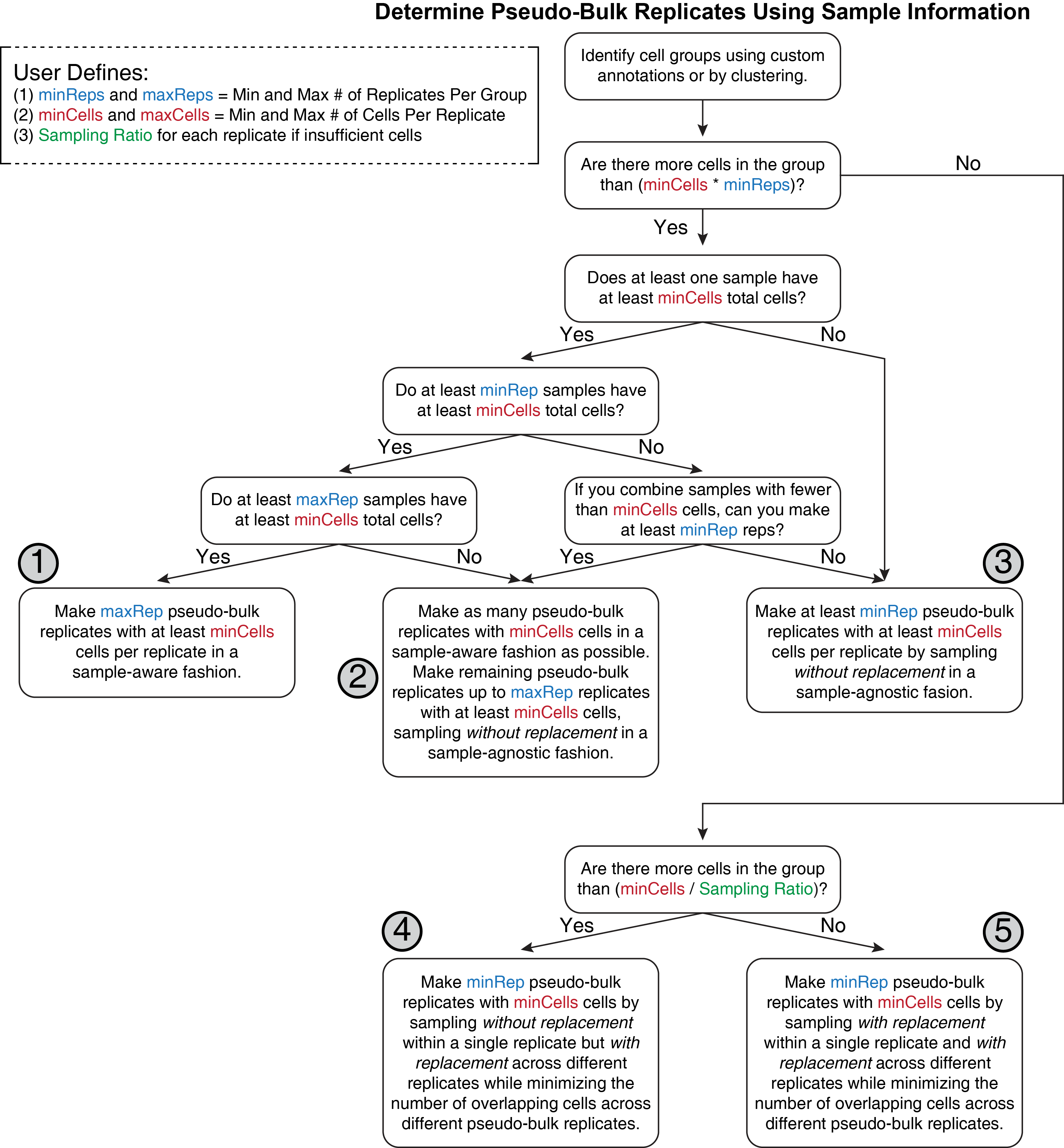
We outline some of the key considerations of this process in words here. First, the user identifies the cell groups to be used - this is often the clusters called by ArchR. Then for each cell grouping, ArchR attempts to create the desired pseudo-bulk replicates. The ideal pseudo-bulk replicate would consist of a sufficient number of cells from a single sample. This maintains sample diversity and biological variation between the replicates. This is what ArchR strives to obtain, but in reality there are 5 possible outcomes in this process, ranked below by preference in ArchR:
- Enough different samples (at least the max # replicates) each have more than the minimum number of cells to create pseudo-bulk replicates in a sample-aware fashion, combining only cells from the same sample into a single replicate.
- Some samples each have more than the minimum number of cells to create pseudo-bulk replicates in a sample-aware fashion. The remaing required replicates are created by combining cells without replacement from samples that are not already represented in the sample-aware pseudobulks.
- No samples have more than the minimum number of cells to create a sample-aware pseudo-bulk replicate but there are more cells than
minCells * minReps. All required replicates are created by combining cells without replacement from in a sample-agnostic fashion. - The total number of cells within a cell grouping is less than the minimum number of cells multiplied by the minimum number of replicates but greater than the minimum number of cells divided by the sampling ratio. Create the minimum number of replicates by sampling without replacement within a single replicate but with replacement across replicates while minimizing the number of cells present in multiple pseudo-bulk replicates.
- The total number of cells within a cell grouping is less than the minimum number of cells divided by the sampling ratio. This means that we must make replicates by sampling with replacement within a single replicate and across different replicates. This is the worst case scenario and users should be cautious about using these pseudo-bulk replicates downstream. This can be controled in various other ArchR functions using the
minCellsparameter.
To illustrate this process, we will use the following example data set:
Sample Cluster1 Cluster2 Cluster3 Cluster4 Cluster5
A 800 600 900 100 75
B 1000 50 400 150 25
C 600 900 100 200 50
D 1200 500 50 50 25
E 900 100 50 150 50
F 700 200 100 100 25And we will set minRep = 3, maxRep = 5, minCells = 300, maxCells = 1000, and sampleRatio = 0.8.
11.1.1 Cluster1
For Cluster1, we have 6 samples (more than maxRep) that all have more than minCells cells (300 cells). This illustrates Option#1 above and we will make 5 pseudo-bulk replicates in a sample aware fashion like so:
Rep1 = 800 cells from SampleA
Rep2 = 1000 cells from SampleB
Rep3 = 1000 cells from SampleD
Rep4 = 900 cells from SampleE
Rep5 = 700 cells from SampleFThere are two things to note about these replicates: (i) SampleC was left out because we had more than enough samples to make maxRep sample-aware pseudo-bulk replicates and SampleC had the fewest number of cells. (ii) Only 1000 cells were used from SampleD because this is the maxCells value.
11.1.2 Cluster2
For Cluster2, we have 3 samples that all have more than minCells cells and a few additional samples that do not. This illustrates Option#2 above and we will make the following pseudo-bulk replicates:
Rep1 = 600 cells from SampleA
Rep2 = 900 cells from SampleC
Rep3 = 500 cells from SampleD
Rep4 = 350 cells [50 cells from SampleB + 100 from SampleE + 200 from SampleF]In this example, Rep4 gets created in a samaple agnostic fashion by sampling without replacement.
11.1.3 Cluster3
For Cluster3, we only have 2 samples that have more than minCells cells which is less than the required minReps. However, if we combine the cells from the remaining samples, we can make one additional replicate with more than minCells. This gives us a total of 3 pseudo-bulk replicates and represents the situation illustrated by Option#3 above. We will make the following replicates:
Rep1 = 900 cells from SampleA
Rep2 = 400 cells from SampleB
Rep3 = 300 cells [100 cells from SampleC + 50 from SampleD + 50 from SampleE + 100 from SampleF]Similar to Cluster2 above, Cluster3 Rep3 is created in a sample agnostic fashion by sampling without replacement across multiple samples.
11.1.4 Cluster4
For Cluster4, the total number of cells is 750 which is less than minCells * minReps (900 cells). In this case, we do not have sufficient cells to make minReps with at least minCells without some form of sampling with replacement. However, the total cells is still greater than minCells / sampleRatio (375 cells) which means that we only have to sample with replacement across different pseudo-bulk replicates, not within a single replicate. This represents the situation illustrated in Option#4 above and we will therefore make the following replicates:
Rep1 = 300 cells [250 unique cells + 25 cells overlapping Rep2 + 25 cells overlapping Rep3]
Rep2 = 300 cells [250 unique cells + 25 cells overlapping Rep1 + 25 cells overlapping Rep3]
Rep3 = 300 cells [250 unique cells + 25 cells overlapping Rep1 + 25 cells overlapping Rep2]In this case, ArchR will minimize the number of cells that overlap between any two pseudo-bulk replicates.
11.1.5 Cluster5
For Cluster5, the total number of cells is 250 which is less than minCells * minReps (900 cells) and less than minCells / sampleRatio (375 cells). This means that we we will have to sample with replacement within each sample and across different replicates to make pseudo-bulk replicates. This represents the least desirable situation illustrated in Option#5 above and we should therefore be cautious in using these pseudo-bulk replicates in downstream analyses. We will therefore make the following replicates:
Rep1 = 300 cells [250 unique cells + 25 cells overlapping Rep2 + 25 cells overlapping Rep3]
Rep2 = 300 cells [250 unique cells + 25 cells overlapping Rep1 + 25 cells overlapping Rep3]
Rep3 = 300 cells [250 unique cells + 25 cells overlapping Rep1 + 25 cells overlapping Rep2]